Overview
As a biochemistry and structural biology lab, we are interested in understanding biological processes at the molecular level. In particular, we focus on macromolecules involved in post-transcriptional and post-translational modifications.
Projects
1. p97 and the ubiquitin system
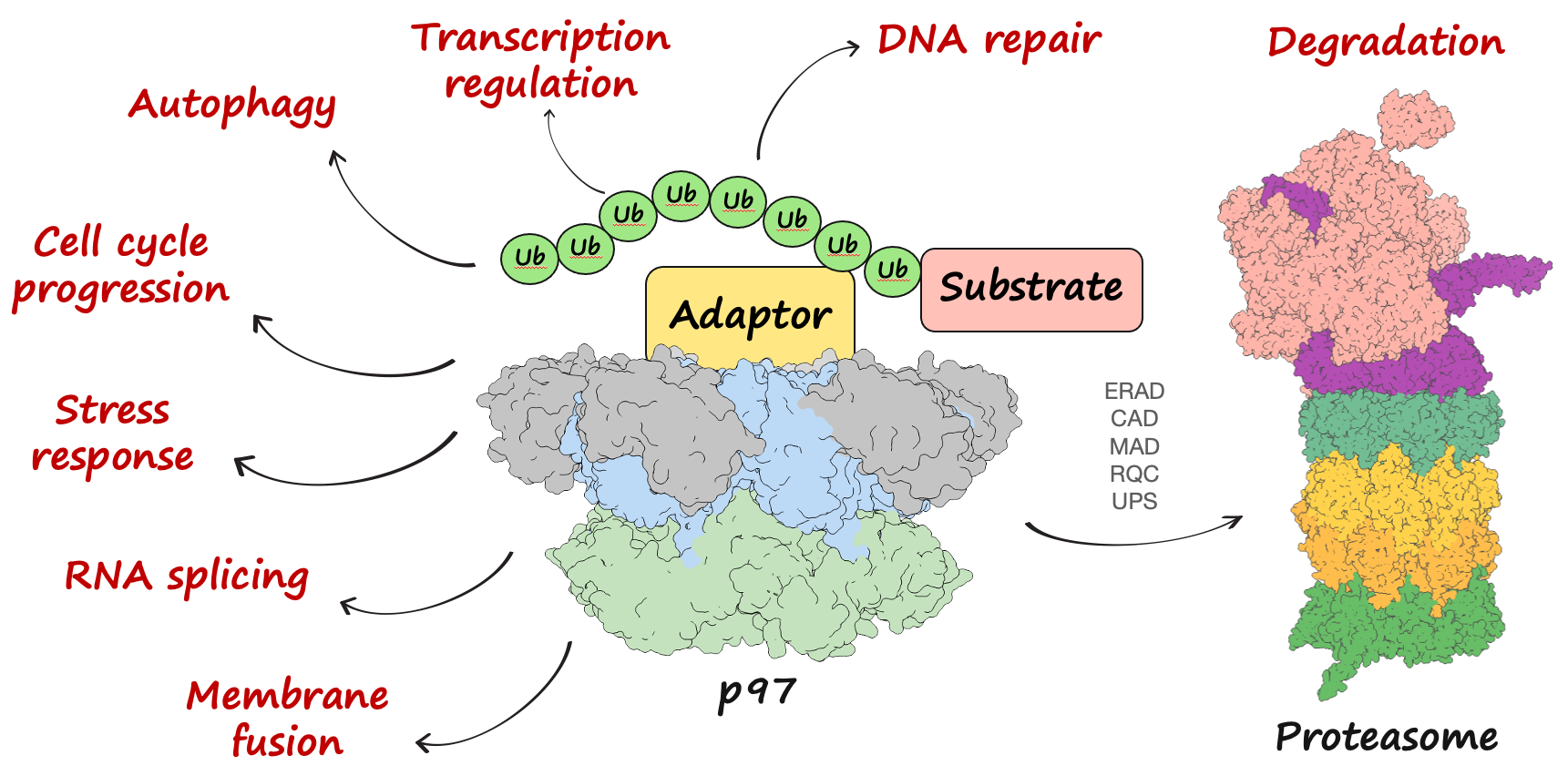 p97 is a central hub of the cellular ubiquitin system. It is highly abundant in cytoplasm and participates in many cellular processes through a variety of cofactors. Yet we know little about how p97 process various ubiquitinated substrates through different cofactors. In addition, disease mutations of p97 have been linked to human neurodegenerative diseases known as multisystem proteinopathy (MSP), but its molecular mechanism remains elusive. On the other hand, the ubiquitin system is extremely diverse, and the functions of atypical polyubiquitin chains are poorly understood. p97 is involved in atypical ubiquitin signaling besides serving as a classical preprocessor for proteasome degradation.
p97 is a central hub of the cellular ubiquitin system. It is highly abundant in cytoplasm and participates in many cellular processes through a variety of cofactors. Yet we know little about how p97 process various ubiquitinated substrates through different cofactors. In addition, disease mutations of p97 have been linked to human neurodegenerative diseases known as multisystem proteinopathy (MSP), but its molecular mechanism remains elusive. On the other hand, the ubiquitin system is extremely diverse, and the functions of atypical polyubiquitin chains are poorly understood. p97 is involved in atypical ubiquitin signaling besides serving as a classical preprocessor for proteasome degradation.
Our recent work made substantial progress on the understanding of translocation mechanism of human p97 and how it is affected by small molecule inhibitors (Pan, et al., Nature Structural & Molecular Biology, 2021, Pan, et al., Nature Communications, 2021). We also developed a synthetic antibody against K29-linked ubiquitin in collaboration with the Kossiakoff lab at UChicago (Yu, et al., Nature Chemical Biology, 2021). In the future, we plan to continue developing linkage-specific antibodies against other ubiquitin chain topologies and use them as tools to further elucidate the function of p97, the mechanism of p97 disease mutations, and the roles played by various cofactors.
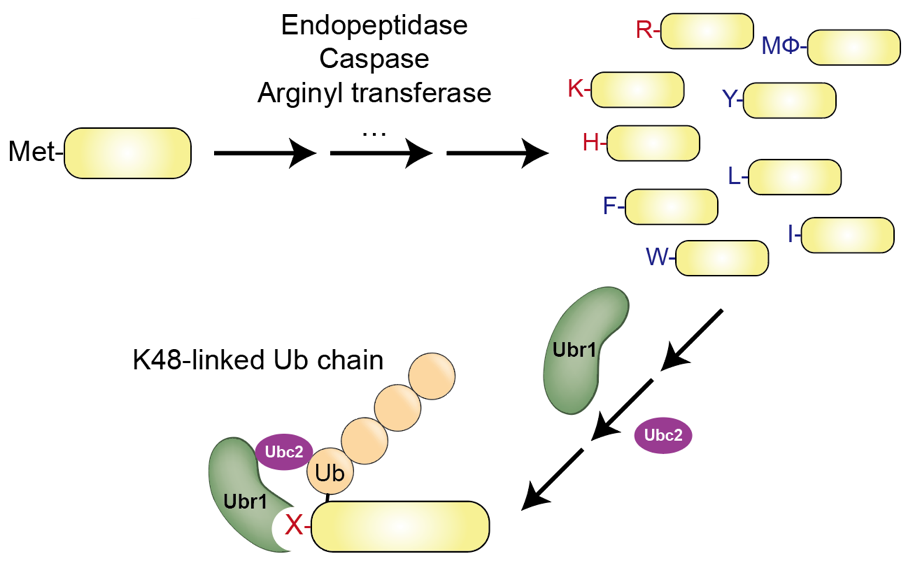 In addition, we have expanded our research to ubiquitin ligases (E3). There are more than 600 E3 ligases/complexes encoded in human proteome. Numerous human diseases including cancers, neurodegenerative diseases, and cardiovascular diseases are related to E3s. Ubr1, an E3 ligase involved in the N-degron pathway, was historically the first E3 to be genetically cloned (Bartel, et al., 1990), but the molecular architecture remained unknown. We have published the first cryoEM structures of yeast Ubr1 in complex with Ubc2 (E2), ubiquitin, and N-degron peptides (Pan, et al., Nature, 2021). We will further explore the human homologs of Ubr1 involved in the N-degron pathway and investigate their potential as therapeutic targets.
In addition, we have expanded our research to ubiquitin ligases (E3). There are more than 600 E3 ligases/complexes encoded in human proteome. Numerous human diseases including cancers, neurodegenerative diseases, and cardiovascular diseases are related to E3s. Ubr1, an E3 ligase involved in the N-degron pathway, was historically the first E3 to be genetically cloned (Bartel, et al., 1990), but the molecular architecture remained unknown. We have published the first cryoEM structures of yeast Ubr1 in complex with Ubc2 (E2), ubiquitin, and N-degron peptides (Pan, et al., Nature, 2021). We will further explore the human homologs of Ubr1 involved in the N-degron pathway and investigate their potential as therapeutic targets.
2. Vault and the ADP-ribosylation system
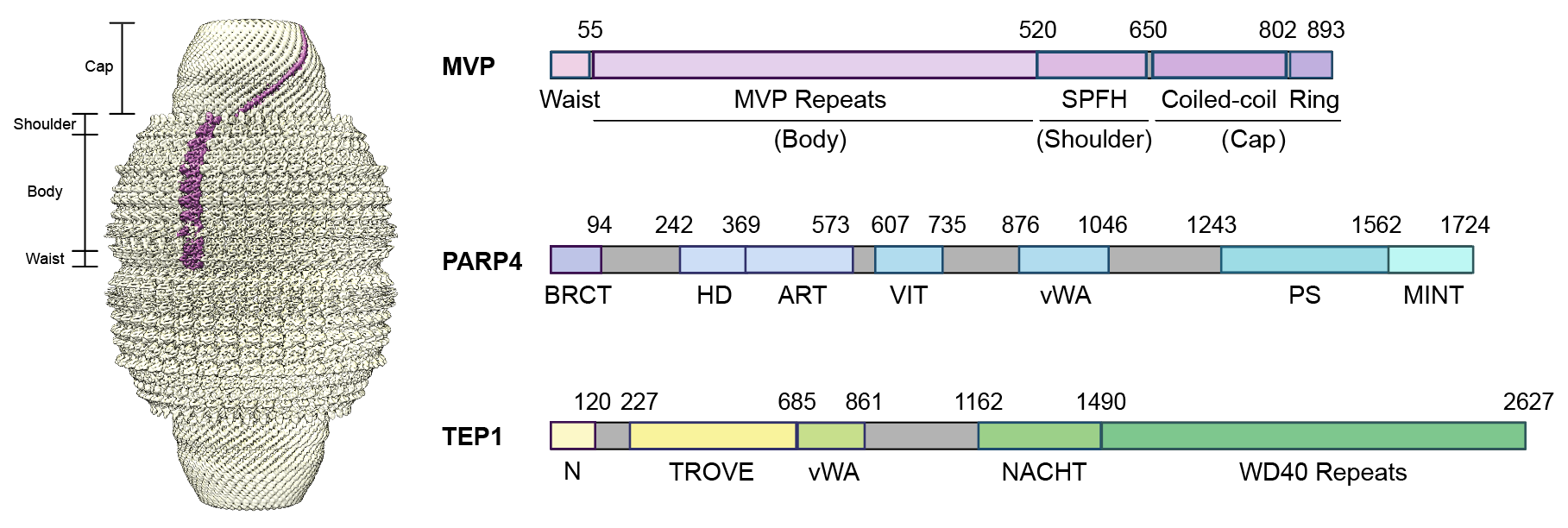 The molecular function of the Vault, the largest protein-RNA complex in eukaryotes, has eluded researchers since its discovery over 30 years ago. The structure of Vault is comprised of 78 copies of the major vault protein (MVP), which coalesce into an oblong cage enclosing at least three “minor” vault components. Although knocking out MVP in healthy mice does not induce a phenotype, MVP knockout mice subjected to lung infection with P. aeruginosa or Influenza A virus showed significantly reduced survival rates compared to their WT counterparts. However, Vault’s mechanism of action in these processes remains unclear. We are focusing on PARP4, the enzyme in the Vault complex catalyzing ADP-ribosylation, a post-translational modification that has been found to facilitate host response against virus infection, and a potential link to the MVP knockout phenotype. Some viruses, including SARS-CoV-2, encode a so-called macrodomain that can reverse the ADP-ribosylation in order to escape this host defense mechanism. Resolving the molecular interactions between PARP4 activity and MVP will therefore shed light on the role of Vault in immune responses. We are using classical structural biology approaches to probe the molecular interactions between PARP4 and MVP. In parallel, we are adopting chemical biology and proteomic approaches to identify the substrates of PARP4. The ultimate goal is to link the function of Vault to its peculiar molecular structure.
The molecular function of the Vault, the largest protein-RNA complex in eukaryotes, has eluded researchers since its discovery over 30 years ago. The structure of Vault is comprised of 78 copies of the major vault protein (MVP), which coalesce into an oblong cage enclosing at least three “minor” vault components. Although knocking out MVP in healthy mice does not induce a phenotype, MVP knockout mice subjected to lung infection with P. aeruginosa or Influenza A virus showed significantly reduced survival rates compared to their WT counterparts. However, Vault’s mechanism of action in these processes remains unclear. We are focusing on PARP4, the enzyme in the Vault complex catalyzing ADP-ribosylation, a post-translational modification that has been found to facilitate host response against virus infection, and a potential link to the MVP knockout phenotype. Some viruses, including SARS-CoV-2, encode a so-called macrodomain that can reverse the ADP-ribosylation in order to escape this host defense mechanism. Resolving the molecular interactions between PARP4 activity and MVP will therefore shed light on the role of Vault in immune responses. We are using classical structural biology approaches to probe the molecular interactions between PARP4 and MVP. In parallel, we are adopting chemical biology and proteomic approaches to identify the substrates of PARP4. The ultimate goal is to link the function of Vault to its peculiar molecular structure.
3. Methyltransferases and proteins involved in mRNA methylation
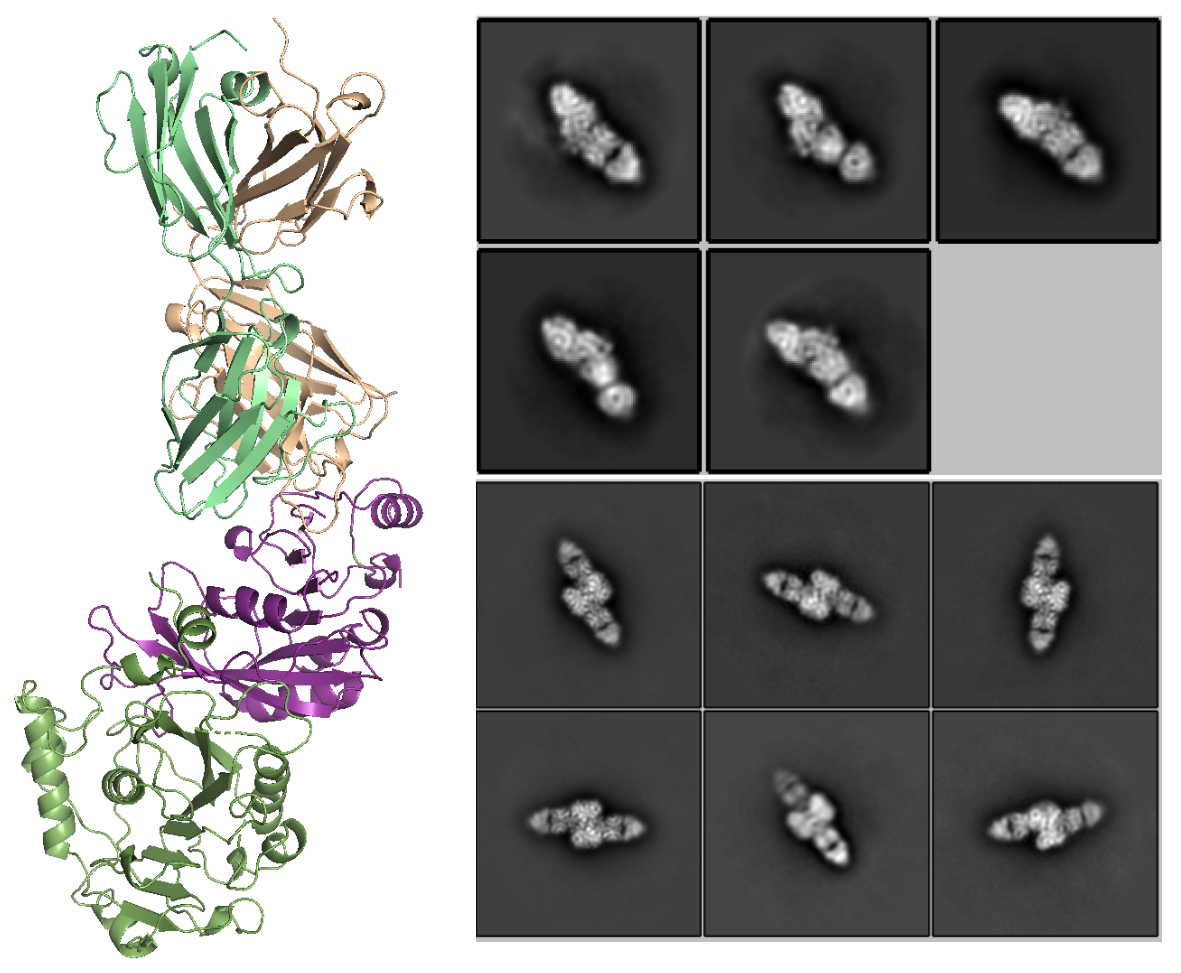 Methylation is an important modification regulating the localization and expression of mRNA. Methyltransferases are the “writer” proteins involved in the modification. Many mutations of methyltransferases are associated with cancers. However, how methyltransferases engage RNA is poorly understood at the atomic level. We are focusing on Mettl3/Mettl14 complex (involved in m6A modification) and taking structural biology and single molecule approaches to address the question of how these methyltransferases achieve substrate specificity. We have developed specific binders for Mettl3/Mettl14 complex (with the Kossiakoff lab) and are conducting single-molecule studies (in collaboration with the Fei lab) to further elucidate the molecular mechanism of Mettl3/Mettl14. Meanwhile, we are collaborating with the He lab and the Fang lab on mechanistic studies of other methyltransferases.
Methylation is an important modification regulating the localization and expression of mRNA. Methyltransferases are the “writer” proteins involved in the modification. Many mutations of methyltransferases are associated with cancers. However, how methyltransferases engage RNA is poorly understood at the atomic level. We are focusing on Mettl3/Mettl14 complex (involved in m6A modification) and taking structural biology and single molecule approaches to address the question of how these methyltransferases achieve substrate specificity. We have developed specific binders for Mettl3/Mettl14 complex (with the Kossiakoff lab) and are conducting single-molecule studies (in collaboration with the Fei lab) to further elucidate the molecular mechanism of Mettl3/Mettl14. Meanwhile, we are collaborating with the He lab and the Fang lab on mechanistic studies of other methyltransferases.
Techniques
Our lab employs two primary structural biology methods: X-ray crystallography and cryo-electron microscopy. Both techniques enable us to resolve atomic structures of macromolecules, providing crucial insights into the mechanisms underlying biological processes and diseases. For more information on these techniques, please visit the resource page. Beyond these core methodologies, we are also exploring the integration of chemical biology, protein engineering, single-molecule techniques, and cell biological approaches to tackle complex and challenging biological questions.
